3️⃣ Predicting early onset dementia - ML Modeling 🚀

The code for this series can be found here 📍
If you prefer to see the contents of the Jupyter notebook, then click here.
All the heavy-lifting has been done… now it’s time to evaluate some ML algorithms.
Evaluating ML Models⌗
The No Free Lunch theorem in Machine Learning stipulates that there is not one algorithm which works perfectly for all dataset. Thus, the performance of differing machine learning classifications will be computed and compared.
The main evaluation metric to gauge algorithm performance is the AUC (Area under ROC curve) score.
The ROC curve displays the trade-off between the true positive rate and the false positive rate.
In the case for diagnosing Dementia, it’s imperative that patients who exhibit symptoms are identified as early as possible (high true positive rate) whilst healthy patients aren’t misdiagnosed with Dementia (low false positive rate) and begin treatment.
AUC is the most appropriate performance measure as it will aid in distinguishing between the two diagnostic groups(Demented/ Nondemented).
Other evaluation metrics are used to compliment the AUC score but don’t carry the same weight. These include:
- Cross-Validation score (or gridsearch score).
- Recall score - The ratio of positive instances that each of our models detect.
- Diagnostic odds ratio (DOR score) is the odds of positivity in individuals with a illness relative to the odds in individuals without an illness.
The
evaluateAlgosfunction callsclassReportwhich computes all of the above evaluation metrics. This function will be called for a series of machine learning algos.
Before evaluation, a global scores table is defined that will record important metrics that are used to evaluate the validity of each model and whether that model is using default parameters or hyperparameters that have been tuned via grid-search or randomized grid seach:
⚠️
A BaselineModel is used as a benchmark model. If algorithms perform better than this benchmark, it reaffirms that applying machine learning techniques to this dataset is applicable.
Please refer to the appendix for some background on each algorithm
Linear classifiers are evaluated using default parameters first:
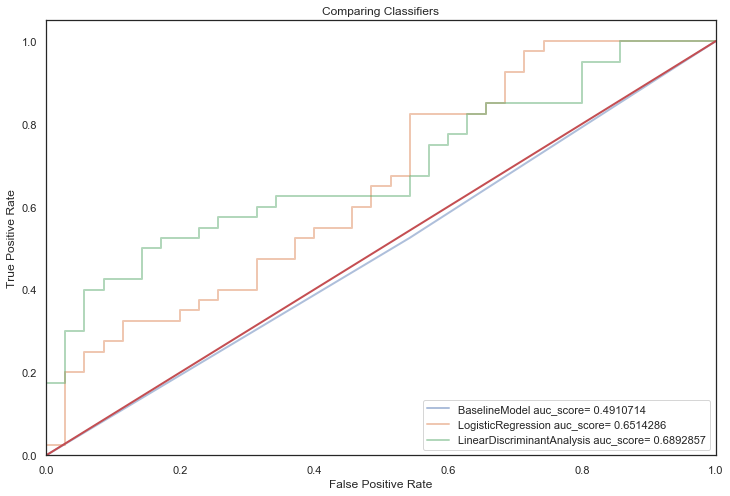
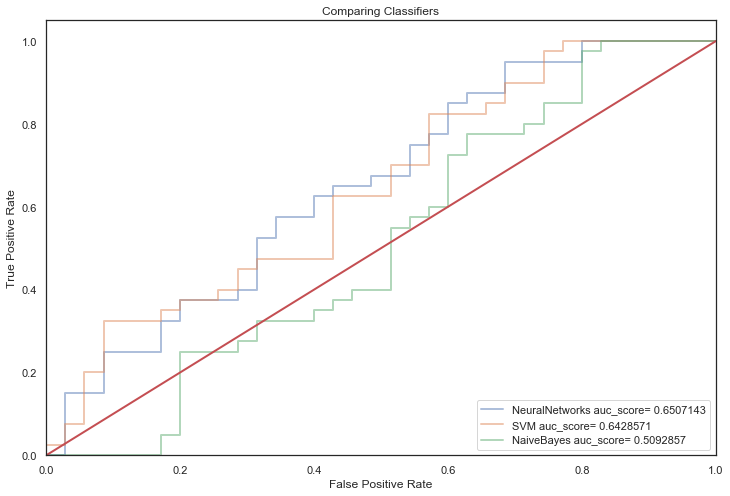
Next, non-linear classifiers with default parameters:
Finally, tree based classifiers:
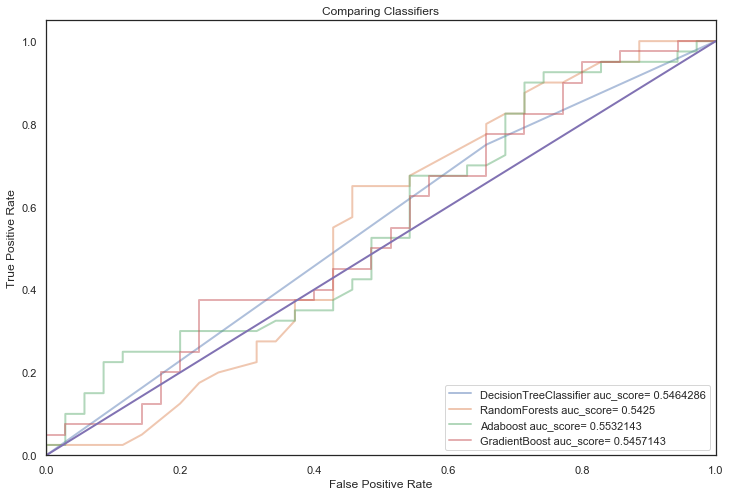
First off, most models with default parameters provided a significant improvement over the baseline model indicating that applying Machine learning techniques to this dataset is applicable.
It is apparent that all models suffer from overfitting, a crux of having a small dataset.
They score strongly in training predictions but generalise poorly on unseen data (overfitting!).
Please refer to follow section in the appendix to see the effects of Overfitting in SVM
The next steps will try and circumvent overfitting by:
Using the Boruta Algorithm to remove irrelevant features and only retain features that fall within an area of absolute acceptance.
Using the GridSearch and RandomizedGridSearch techniques with cross validation to finely tune hyperparameters that could unknowlingly exasperate overfitting.
Part 2️⃣
Feature selection⌗
Convert the arrays back into q tables:
Feature selection is the process of finding a subset of features in the dataset X which have the greatest discriminatory power with respect to the target variable y.
If feature selection is ignored:
- It becomes computationally expensive as the model is processing a large number of features.
- garbage in, garbage out. When the number of features is significantly higher than optimal, a dip in accuracy is observed.
Occam's razorstipulates that a problem should be simplified by removing irrelevant features that would introduce unncessary noise. If a model remembers noise in a small dataset, it could generalise poorly on unseen data.
Ideally, instead of manually going through each feature to decide if any relationship exists between it and the target, an algorithm that is able to autonomously decide whether any given feature of X bears some predictive value about y is desired.
This is what the Boruta algorithm does.
The iteration count for the Boruta algorithm is arbitrary. The user provides a list of integers where:
- The iteration count is equal to the length of the list
- Each value of the list is used as a random seed value
So in the below case, 80 runs are executed against the training dataset X_train with the random seed value iterating each run (starting at 1, finishing at 80). The user decides how many features they extract from the area of acceptance (3 in this case):
Important features [nwbv mmse educ] are extracted and kept using the Boruta Algorithm.
The remaining features are dropped:
Train and test sets are converted back to python arrays:
Hyperparameter Tuning⌗
In order to further improve the AUC score for each model, the hyperparameters for each classifier are optimized using one of the following techniques:
- GridSearch
- RandomizedSearchCV
GridSearch simplifies the process of implementing and optimizing hyperparameters. By passing a dictionary of hyperparameters to be tested to the GridSearchCV function, all possible combinations of these values can be evaluated using cross-validation on a model selected by the user.
GridSearch is useful when the hyperparameter combinations to be explored are limited.
However, when the hyperparameter space is vast, it is better to use RandomizedSearchCV. This method operates similarly to GridSearch but with a key difference - rather than testing all possible hyperparameter combinations,
RandomizedSearchCVwill assess a fixed number of hyperparameter sets drawn from specified probability distributions. It is usually preferred over GridSearch as the user has more control over the computational resources by setting the number of iterations.
An optimalModels key table is defined that will tabulate the optimal parameters found using the grid-search/randomized grid-search technique for a particular classifier.
An optimal model will then be used by a web application to predict whether an individual is displaying alzheimer symptoms.
A series of dictionaries are defined that represent the parameter space for each algorithm:
A dictionary is defined to map each parameter space to its algorithm:
The parameter space for each algorithm is hypertuned to compute optimal parameters:
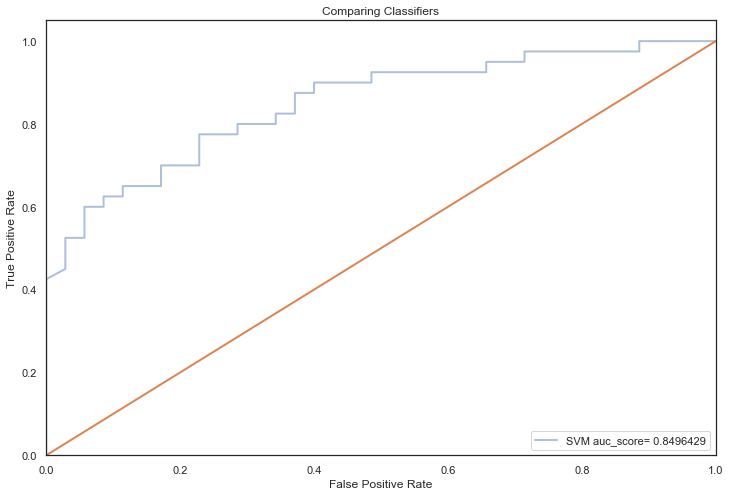
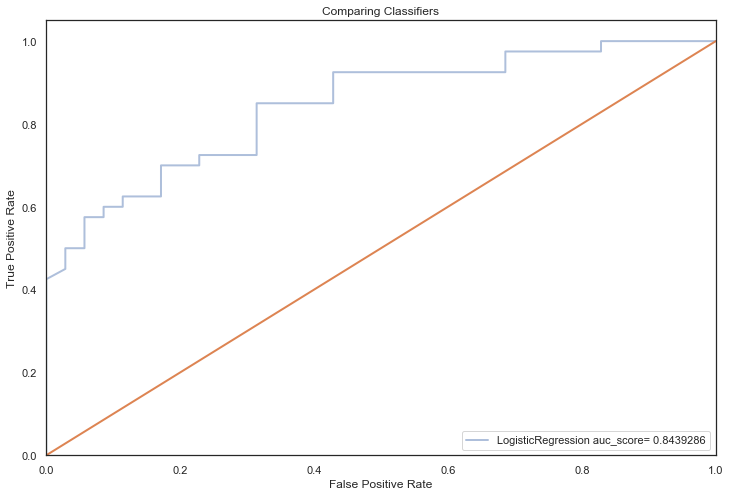
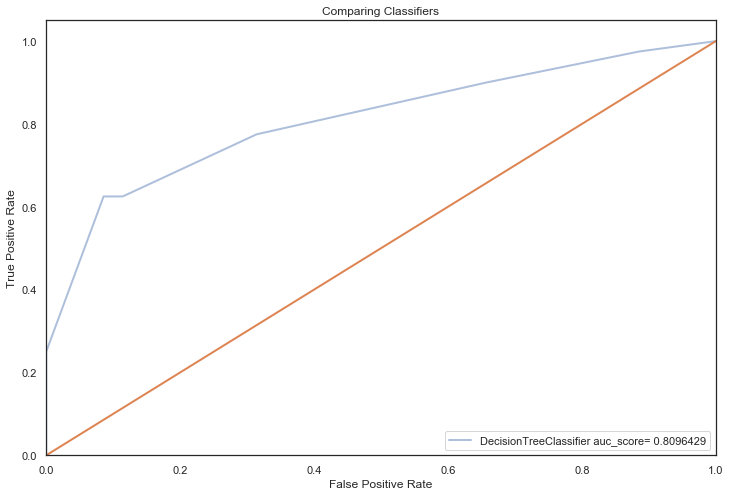
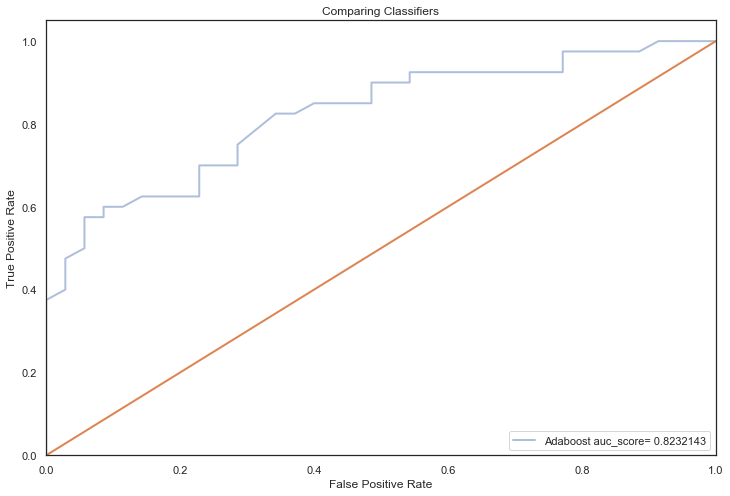
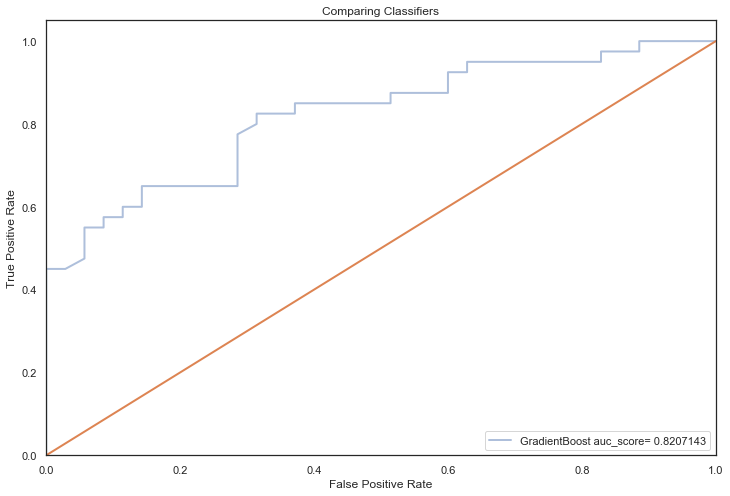
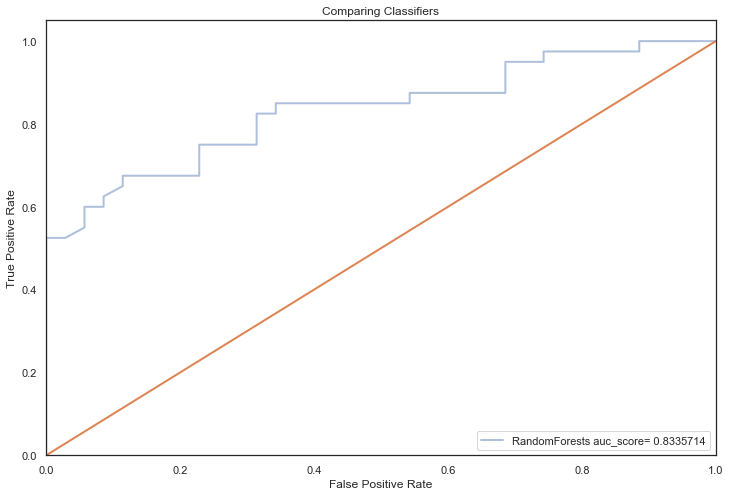
All models hypertuned via RandomizedGridSearchCV computed higher AUC scores than the previous no tuning evaluation, an indication that hyperparameter tuning via RandomizedSearchCV coupled with feature selection techniques, improved performance significantly (the closer to 1, the better):
Conclusion⌗
The model with the highest AUC score is the SVM algo 🎉
The SVM model is relatively simple compared to boosting and ensemble algorithms, which suggest that these results support the principle of Occam’s razor, where for a small dataset, the use of straightforward models with minimal assumptions often leads to the best results.
Although, there is still some overfitting happening (training acc > test acc), mainly due to the size of this dataset, it is not as consequential as previous, as the training accuracies have decreased significantly whilst the test accuracies have risen substantially.
In essence, the models aren’t learning as many particulars in the training dataset and therefore generalising better on unseen data.
Previously, models were learning details and noise in the training data to the extent that it was generalising very poorly on unseen data.